Sourcing Data from S3 with Drake
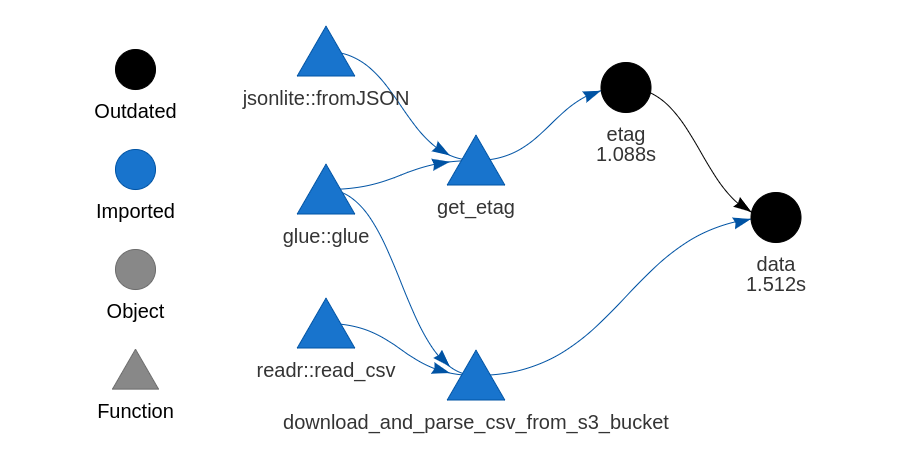
drake is a package for orchestrating R workflows. Suppose I have some data in S3 that I want to pull into R through a drake plan. In this post I’ll use the S3 object’s ETag to make drake only re-download the data if it’s changed.
This covers the scenario in which the object name in S3 stays the same. If I had, say, data being uploaded each day with an object name suffixed with the date, then I wouldn’t bother checking for any changes.
Connecting to S3
Both the aws.s3 package and the PAWS package will connect to S3 from R. I’ve used both of these packages, and there’s nothing wrong with them, but I always find myself going back to wrapping AWS CLI commands. I’m not saying this is the best way to use AWS from within R, but it works, although I haven’t tested this on anything other than Linux.
By this point I’ve run aws configure in a terminal to make sure that I can actually connect to AWS. I’ve also created an S3 bucket.
There are two ways to connect to S3 from the AWS CLI. s3 commands are more high-level than s3api commands, but I’ll need to use both here.
Uploading some data
I’ll start by uploading some CSV data to my bucket using an s3 command, so that I have something to source in my drake plan. What I really like about the s3 commands is that I don’t have to mess around with any multi-part uploads, as the AWS CLI takes care of all that complexity for me.
I’ll create a function that forms and executes the command. My command needs to be of the form aws s3 cp $SOURCE $TARGET. The $SOURCE or $TARGET variables can be either local files or objects on S3, with objects prefixed with “s3://$BUCKET”. My function will take a data frame and, using the name of that data frame, determine the path of the object on S3. A more sophisticated function would be more flexible about how I’m storing the data, but this will do for my demonstration.
Note the use of shQuote here, a base function that quotes a string to be passed to a shell.
upload_data_to_s3_bucket_as_csv <- function(data, bucket) {
object_name <- paste0(deparse(substitute(data)), ".csv")
temp_file <- tempfile()
# delete this temp file afterwards, even if this function errors
on.exit(unlink(temp_file))
readr::write_csv(data, temp_file)
quoted_file_path <- shQuote(temp_file)
quoted_object_path <- shQuote(glue::glue("s3://{bucket}/{object_name}"))
system(glue::glue("aws s3 cp {quoted_file_path} {quoted_object_path}"))
}Getting object metadata
The ETag is a hash that changes when the object changes1. It’s a short string like “de3b6f4731f18de03e51a5fea8102c93”. No matter how big an object is, the ETag stays the same size, and is quick to retrieve. This means that we can check the ETag every time a drake plan is made without spending too much time, and only re-download the actual data if drake detects a change in this value.
I need to use a lower-level s3api command here. The head-object command retrieves object metadata. I convert that metadata from JSON, extract the ETag, and remove the stray quotation marks around it.
get_etag <- function(object, bucket) {
response <- system(
glue::glue("aws s3api head-object --bucket {bucket} --key {object}"),
intern = TRUE
)
raw_etag <- jsonlite::fromJSON(response)$ETag
gsub("\"", "", raw_etag)
}Downloading from S3
I’ll once again use an s3 command to download data from an S3. This function is very similar to the upload function, with the source and target reversed.
download_and_parse_csv_from_s3_bucket <- function(object, bucket) {
temp_file <- tempfile()
# delete this temp file afterwards, even if this function errors
on.exit(unlink(temp_file))
quoted_file_path <- shQuote(temp_file)
quoted_object_path <- shQuote(glue::glue("s3://{bucket}/{object}"))
system(glue::glue("aws s3 cp {quoted_object_path} {quoted_file_path}"))
readr::read_csv(temp_file)
}Generating some random data
I’ll need some data to upload to my bucket and then retrieve. Here’s my go-to function for generating a data frame of random bits, adapated from this StackOverflow answer:
generate_random_data <- function(nrow = 1000, ncol = 10) {
data.frame(replicate(ncol, sample(0:1, nrow, rep = TRUE)))
}Now I’ll upload some random data to my bucket. I’ve created a bucket “ocelittle”, which is the unofficial name of ocelot kittens. This has nothing to do with AWS; I just needed a unique name for the bucket.
some_random_data <- generate_random_data()
upload_data_to_s3_bucket_as_csv(some_random_data, bucket = "ocelittle")
get_etag("some_random_data.csv", bucket = "ocelittle")
#> [1] "104b796e58ef578339253f1f04673388"Method 1: A separate target for the ETag
There are two equally valid ways to structure the drake plan to check the ETag. They’re effectively equivalent, but there’s some slight variation in how the targets are displayed when I run drake::vis_drake_graph.
In this first method, I’ll create a separate target for the ETag so that it appears in my drake plan visualisations, as in the plot at the top of this page. Pay close attention to the conditions for each trigger:
s3_plan <- drake::drake_plan(
etag = target(
get_etag("some_random_data.csv", "ocelittle"),
trigger = trigger(condition = TRUE)
),
data = target(
download_and_parse_csv_from_s3_bucket("some_random_data.csv", "ocelittle"),
trigger = trigger(change = etag)
)
)The condition for the etag target is TRUE, which means that this target will always run when I make the drake plan. The data target only runs when the value of the etag target has changed. When I make this plan for the first time, both targets are executed:
drake::make(s3_plan)
#> ▶ target etag
#> ▶ target data
#> Parsed with column specification:
#> cols(
#> X1 = col_double(),
#> X2 = col_double(),
#> X3 = col_double(),
#> X4 = col_double(),
#> X5 = col_double(),
#> X6 = col_double(),
#> X7 = col_double(),
#> X8 = col_double(),
#> X9 = col_double(),
#> X10 = col_double()
#> )When I run the plan a second time, the etag target runs, as expected. But as the object’s ETag hasn’t changed, drake doesn’t execute the data target.
drake::make(s3_plan)
#> ▶ target etagNow I’ll generate some new random data, and overwrite the previous CSV:
some_random_data <- generate_random_data()
upload_data_to_s3_bucket_as_csv(some_random_data, bucket = "ocelittle")
get_etag("some_random_data.csv", bucket = "ocelittle")
#> [1] "5b978808807d201824757e7b703d8910"drake detects the change and re-downloads the data:
drake::make(s3_plan)
#> ▶ target etag
#> ▶ target data
#> Parsed with column specification:
#> cols(
#> X1 = col_double(),
#> X2 = col_double(),
#> X3 = col_double(),
#> X4 = col_double(),
#> X5 = col_double(),
#> X6 = col_double(),
#> X7 = col_double(),
#> X8 = col_double(),
#> X9 = col_double(),
#> X10 = col_double()
#> )Method 2: Embedding the ETag in the data target
Rather than having a separate target for the etag, I can use put the get_etag function directly into the change condition for the data download target. This won’t show the ETag when I run drake::drake_vis_graph.
First, I’ll clean the drake cache:
drake::clean()The change trigger accepts any R expression, so it accepts the get_etag function. This will run every time the plan is made.
s3_plan_2 <- drake::drake_plan(
data = target(
download_and_parse_csv_from_s3_bucket("some_random_data.csv", "ocelittle"),
trigger = trigger(change = get_etag("some_random_data.csv", "ocelittle"))
)
)drake::make(s3_plan_2)
#> ▶ target data
#> Parsed with column specification:
#> cols(
#> X1 = col_double(),
#> X2 = col_double(),
#> X3 = col_double(),
#> X4 = col_double(),
#> X5 = col_double(),
#> X6 = col_double(),
#> X7 = col_double(),
#> X8 = col_double(),
#> X9 = col_double(),
#> X10 = col_double()
#> )drake::make(s3_plan_2)
#> ✔ All targets are already up to date.And now, just to check, I’ll upload some new data and make sure that drake downloads it:
some_random_data <- generate_random_data()
upload_data_to_s3_bucket_as_csv(some_random_data, bucket = "ocelittle")
get_etag("some_random_data.csv", bucket = "ocelittle")
#> [1] "afce4300c7ea473c81b5a9f0f9712af3"drake::make(s3_plan_2)
#> ▶ target data
#> Parsed with column specification:
#> cols(
#> X1 = col_double(),
#> X2 = col_double(),
#> X3 = col_double(),
#> X4 = col_double(),
#> X5 = col_double(),
#> X6 = col_double(),
#> X7 = col_double(),
#> X8 = col_double(),
#> X9 = col_double(),
#> X10 = col_double()
#> )Once again, drake detects the change and re-downloads the data.
devtools::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.0.0 (2020-04-24)
#> os Ubuntu 20.04.1 LTS
#> system x86_64, linux-gnu
#> ui X11
#> language en_AU:en
#> collate en_AU.UTF-8
#> ctype en_AU.UTF-8
#> tz Australia/Melbourne
#> date 2020-08-23
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date lib source
#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.0)
#> backports 1.1.8 2020-06-17 [1] CRAN (R 4.0.0)
#> base64url 1.4 2018-05-14 [1] CRAN (R 4.0.0)
#> callr 3.4.3 2020-03-28 [1] CRAN (R 4.0.0)
#> cli 2.0.2 2020-02-28 [1] CRAN (R 4.0.0)
#> crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.0)
#> desc 1.2.0 2018-05-01 [1] CRAN (R 4.0.0)
#> devtools 2.3.0 2020-04-10 [1] CRAN (R 4.0.0)
#> digest 0.6.25 2020-02-23 [1] CRAN (R 4.0.0)
#> downlit 0.0.0.9000 2020-07-25 [1] Github (r-lib/downlit@ed969d0)
#> drake * 7.12.4 2020-07-30 [1] Github (ropensci/drake@20cd701)
#> ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.0)
#> evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.0)
#> fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.0)
#> filelock 1.0.2 2018-10-05 [1] CRAN (R 4.0.0)
#> fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.0)
#> glue 1.4.1 2020-05-13 [1] CRAN (R 4.0.0)
#> here 0.1 2017-05-28 [1] CRAN (R 4.0.0)
#> hms 0.5.3 2020-01-08 [1] CRAN (R 4.0.0)
#> htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.0)
#> hugodown 0.0.0.9000 2020-08-13 [1] Github (r-lib/hugodown@2af491d)
#> igraph 1.2.5 2020-03-19 [1] CRAN (R 4.0.0)
#> jsonlite 1.7.0 2020-06-25 [1] CRAN (R 4.0.0)
#> knitr 1.29 2020-06-23 [1] CRAN (R 4.0.0)
#> lattice 0.20-41 2020-04-02 [4] CRAN (R 4.0.0)
#> lifecycle 0.2.0 2020-03-06 [1] CRAN (R 4.0.0)
#> magrittr 1.5 2014-11-22 [1] CRAN (R 4.0.0)
#> Matrix 1.2-18 2019-11-27 [4] CRAN (R 4.0.0)
#> memoise 1.1.0.9000 2020-05-09 [1] Github (hadley/memoise@4aefd9f)
#> pillar 1.4.6 2020-07-10 [1] CRAN (R 4.0.0)
#> pkgbuild 1.1.0 2020-07-13 [1] CRAN (R 4.0.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.0)
#> pkgload 1.1.0 2020-05-29 [1] CRAN (R 4.0.0)
#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.0.0)
#> processx 3.4.3 2020-07-05 [1] CRAN (R 4.0.0)
#> progress 1.2.2 2019-05-16 [1] CRAN (R 4.0.0)
#> ps 1.3.4 2020-08-11 [1] CRAN (R 4.0.0)
#> purrr 0.3.4 2020-04-17 [1] CRAN (R 4.0.0)
#> R6 2.4.1 2019-11-12 [1] CRAN (R 4.0.0)
#> Rcpp 1.0.5 2020-07-06 [1] CRAN (R 4.0.0)
#> readr 1.3.1 2018-12-21 [1] CRAN (R 4.0.0)
#> remotes 2.1.1 2020-02-15 [1] CRAN (R 4.0.0)
#> reticulate 1.16 2020-05-27 [1] CRAN (R 4.0.0)
#> rlang 0.4.7 2020-07-09 [1] CRAN (R 4.0.0)
#> rmarkdown 2.3.3 2020-08-13 [1] Github (rstudio/rmarkdown@204aa41)
#> rprojroot 1.3-2 2018-01-03 [1] CRAN (R 4.0.0)
#> sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.0)
#> storr 1.2.1 2018-10-18 [1] CRAN (R 4.0.0)
#> stringi 1.4.6 2020-02-17 [1] CRAN (R 4.0.0)
#> stringr 1.4.0 2019-02-10 [1] CRAN (R 4.0.0)
#> testthat 2.3.2 2020-03-02 [1] CRAN (R 4.0.0)
#> tibble 3.0.3 2020-07-10 [1] CRAN (R 4.0.0)
#> tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.0)
#> txtq 0.2.3 2020-06-23 [1] CRAN (R 4.0.0)
#> usethis 1.6.1 2020-04-29 [1] CRAN (R 4.0.0)
#> vctrs 0.3.2 2020-07-15 [1] CRAN (R 4.0.0)
#> withr 2.2.0 2020-04-20 [1] CRAN (R 4.0.0)
#> xfun 0.16 2020-07-24 [1] CRAN (R 4.0.0)
#> yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.0)
#>
#> [1] /home/mdneuzerling/R/x86_64-pc-linux-gnu-library/4.0
#> [2] /usr/local/lib/R/site-library
#> [3] /usr/lib/R/site-library
#> [4] /usr/lib/R/library
